Chirality Check
In chemistry, a molecule is chiral if it cannot be superimposed onto its mirror image by any combination of translation and rotation. These non-superposable mirror images are called enantiomers which share identical chemical and physical properties, but have distinct chemical reactivity and optical rotation properties. Checking whether two structures are enantiomers can be formulated as a Procrustes problem.
Rotational Procrustes
Given matrix \(\mathbf{A}_{m \times n}\) and a reference \(\mathbf{B}_{m \times n}\), find the rotational transformation matrix \(\mathbf{R}_{n \times n}\) that makes \(\mathbf{A}\) as close as possible to \(\mathbf{B}\), i.e.,
\begin{equation} \underbrace{\min}_{\left\{\mathbf{R} \left| {\mathbf{R}^{-1} = {\mathbf{R}}^\dagger \atop \left| \mathbf{R} \right| = 1} \right. \right\}} \|\mathbf{A}\mathbf{R} - \mathbf{B}\|_{F}^2 \end{equation}
Orthogonal Procrustes
Given matrix \(\mathbf{A}_{m \times n}\) and a reference \(\mathbf{B}_{m \times n}\), find the find the orthogonal transformation matrix \(\mathbf{Q}_{n \times n}\) that makes \(\mathbf{A}\) as close as possible to \(\mathbf{B}\), i.e.,
\begin{equation} \underbrace{\min}_{\left\{\mathbf{Q} | \mathbf{Q}^{-1} = {\mathbf{Q}}^\dagger \right\}} \|\mathbf{A}\mathbf{Q} - \mathbf{B}\|_{F}^2 \end{equation}
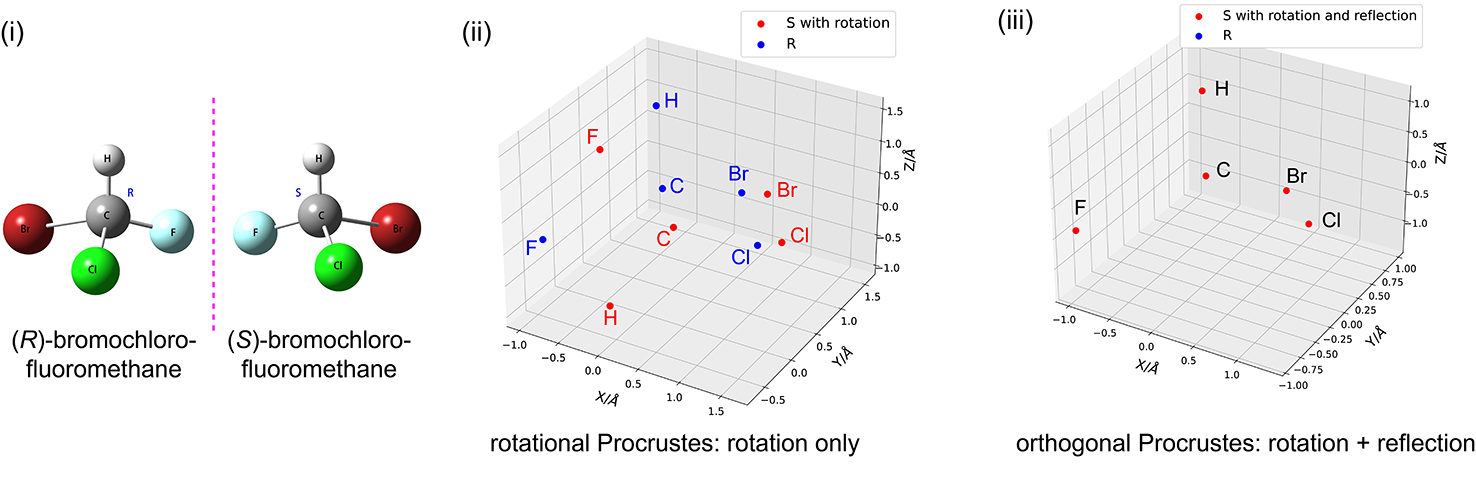
In the code block below, we use the procrustes library to check whether two geometries of the CHFClBr molecule are enantiomers; see Fig. (i). The 3D-Cartesian coordinates of molecules are loaded from their XYZ files using IOData library. Testing whether the coordinates can be matched through translation and rotation (i.e., rotational Procrustes) reveals that these two structures are not identical; see Fig (ii). However, the two coordinates are
enantiomers because they can be matched through translation, rotation, and reflection (i.e., orthogonal Procrustes) as shown in Fig (iii).
Download IOData & Matplotlib Libraries & Example Files
Install IOData Package and Matplotlib, if there are not available on your system; this is required on Binder.
Download enantiomer1.xyz and enantiomer2.xyz files used in the example below which are stored in Procrustes GitHub repository example files.
[ ]:
# If needed, install IOData library (this is required on Binder)
import sys
# See https://jakevdp.github.io/blog/2017/12/05/installing-python-packages-from-jupyter/
!{sys.executable} -m pip install qc-iodata
!{sys.executable} -m pip install matplotlib
[ ]:
# If needed, download the example files
import os
import urllib.request
fpath = "notebook_data/chirality_checking/"
if not os.path.exists(fpath):
os.makedirs(fpath, exist_ok=True)
urllib.request.urlretrieve(
"https://raw.githubusercontent.com/theochem/procrustes/master/doc/notebooks/notebook_data/chirality_checking/enantiomer1.xyz",
os.path.join(fpath, "enantiomer1.xyz")
)
urllib.request.urlretrieve(
"https://raw.githubusercontent.com/theochem/procrustes/master/doc/notebooks/notebook_data/chirality_checking/enantiomer2.xyz",
os.path.join(fpath, "enantiomer2.xyz")
)
Procrustes Analysis
[1]:
# chirality check with rotational and orthogonal Procrustes
from pathlib import Path
import numpy as np
from iodata import load_one
from procrustes import orthogonal, rotational
# load CHClFBr enantiomers' coordinates from XYZ files
a = load_one(Path("notebook_data/chirality_checking/enantiomer1.xyz")).atcoords
b = load_one(Path("notebook_data/chirality_checking/enantiomer2.xyz")).atcoords
# rotational Procrustes on a & b coordinates
result_rot = rotational(a, b, translate=True, scale=False)
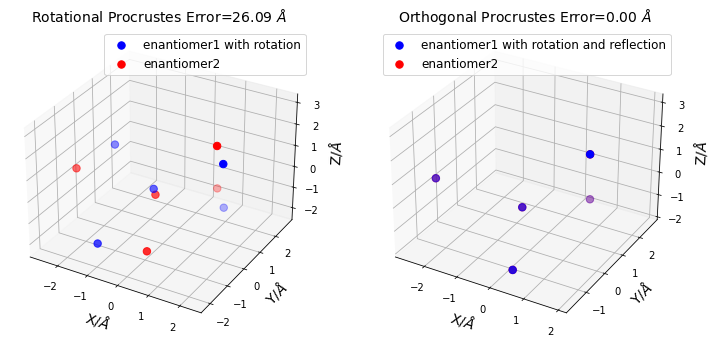
print("Rotational Procrustes Error = ", result_rot.error) # output: 26.085545
# orthogonal Procrustes on a & b coordinates
result_ortho = orthogonal(a, b, translate=True, scale=False)
print("Orthogonal Procrustes Error = ", result_ortho.error) # output: 4.432878e-08
Rotational Procrustes Error = 26.08554575402178
Orthogonal Procrustes Error = 4.432878638501341e-08
Plot Procrustes Results
[2]:
# Plot outputs of Procrustes
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(12, 10))
# =============
# First Subplot
# =============
# set up the axes for the first plot
ax = fig.add_subplot(1, 2, 1, projection='3d')
# coordinates of rotated molecule A and molecule B
a_rot = np.dot(result_rot.new_a, result_rot.t)
coords1, coords2 = a_rot, result_rot.new_b
title = "Rotational Procrustes Error={:0.2f} $\AA$".format(result_rot.error)
ax.scatter(xs=coords1[:, 0], ys=coords1[:, 1], zs=coords1[:, 2],
marker="o", color="blue", s=55, label="enantiomer1 with rotation")
ax.scatter(xs=coords2[:, 0], ys=coords2[:, 1], zs=coords2[:, 2],
marker="o", color="red", s=55, label="enantiomer2")
ax.set_xlabel("X/$\AA$", fontsize=14)
ax.set_ylabel("Y/$\AA$", fontsize=14)
ax.set_zlabel("Z/$\AA$", fontsize=14)
ax.legend(fontsize=12, loc="best")
plt.title(title, fontsize=14)
# ==============
# Second Subplot
# ==============
# set up the axes for the second plot
ax = fig.add_subplot(1, 2, 2, projection='3d')
# coordinates of rotated-and-refelcted molecule A and molecule B
a_rot = np.dot(result_ortho.new_a, result_ortho.t)
coords1, coords2 = a_rot, result_rot.new_b
title="Orthogonal Procrustes Error={:0.2f} $\AA$".format(result_ortho.error)
ax.scatter(xs=coords1[:, 0], ys=coords1[:, 1], zs=coords1[:, 2],
marker="o", color="blue", s=55, label="enantiomer1 with rotation and reflection")
ax.scatter(xs=coords2[:, 0], ys=coords2[:, 1], zs=coords2[:, 2],
marker="o", color="red", s=55, label="enantiomer2")
ax.set_xlabel("X/$\AA$", fontsize=14)
ax.set_ylabel("Y/$\AA$", fontsize=14)
ax.set_zlabel("Z/$\AA$", fontsize=14)
ax.legend(fontsize=12, loc="best")
plt.title(title, fontsize=14)
plt.show()
The corresponding file can be obtained from:
Jupyter Notebook:
Chirality_Check.ipynb